Introduction
So far in my data science journey I have mostly met reasonably small datasets, i.e. they always fit in my machine’s memory without any issue, so I was able to handle them without even having to think about enhancing my code for performance.
Recently I was working on a pet project aiming at performing customers segmentation on a dataset from a major sport retailer. The data contained 8+ million rows and whilst it still fit in memory, I soon noticed that performing exploratory data analysis was becoming too slow to allow me experiment freely.
Therefore I had to find ways to optimize my code, and I found two great ways to do so:
- leverage vectorized operations
- use cython, i.e. C-extension for python
Details about both options are described below.
But first let’s say a few words about profiling
Profiling
Profiling is a way of finding out how much resources/time is required for your code to run.
It’s useful to either compare two pieces of code that perform identical tasks in order to identify which one runs the fastest, or to identify bottlenecks in your code that you can then improve to speed up execution.
Jupyter magic %timeit
The simplest profiling tool comes with jupyter notebook, it’s a magic called %timeit.
Here is a simple use case. Let’s find out how long it takes to sort one million integers randomly picked between 0 and 999
%timeit np.sort(np.random.randint(0,1000,10**6))
#10 loops, best of 3: 58.6 ms per loop
How to understand this? The timeit magic performed 3 times in a row 10 runs of the comand. It found out that for the fastest of the 3 runs, the command took 58.6 ms to execute in average (so it took the total 10 runs time and divided it by 10).
Why 3 times?
Because your computer might be busy doing something else that will impact the command performance. Therefore you want to reduce bias by executing the command a few times and take the best of it.
Why 10 loops?
If your command it very simple and extremly fast to execute, the timing measure won’t be very precise. In that case, better run 10 or 100 loops and output the average, this provides better accurary. Timeit chooses by itself how many loops to perform based on the command complexity.
For instance if my list contains only 10 000 integers instead of one million, the sort is faster, therefore loops of 100 instead of 10 are executed:
%timeit np.sort(np.random.randint(0,1000,10**5))
#100 loops, best of 3: 5.51 ms per loop
These parameters can be configured (‘-r’ specified the number of runs, ‘-n’ the number of loops), see the timeit manual for more.
Also here i want to hightlight that we didn’t only calculate the sorting time, but the list generation time, since both were inculded in the same command. So if we want to understand exactly the performance of the sort algorithm, then we need to move the list generation out of the command which we profile.
listy = np.random.randint(0,1000,10**5)
%timeit np.sort(listy)
#100 loops, best of 3: 5.16 ms per loop
As expected, we now get a faster result! The main learning is that we should be careful of what exactly we are measuing !
The cProfile package
The cProfike package provides more advanced features for profiling. It actually gives details about which part of your code is consumming most resources.
Getting back to our random list generation and sorting, this is what we obtain:
import cProfile
cProfile.run('np.sort(np.random.randint(0,1000,10**6))')
9 function calls in 0.082 seconds
Ordered by: standard name
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.082 0.082 <string>:1(<module>)
1 0.000 0.000 0.062 0.062 fromnumeric.py:717(sort)
1 0.000 0.000 0.000 0.000 numeric.py:484(asanyarray)
1 0.000 0.000 0.082 0.082 {built-in method builtins.exec}
1 0.000 0.000 0.000 0.000 {built-in method numpy.core.multiarray.array}
1 0.004 0.004 0.004 0.004 {method 'copy' of 'numpy.ndarray' objects}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
1 0.020 0.020 0.020 0.020 {method 'randint' of 'mtrand.RandomState' objects}
1 0.058 0.058 0.058 0.058 {method 'sort' of 'numpy.ndarray' objects}
The tottime column shows that the randint command took 20 milliseconds to run, and the sort 58.
With these tools at hand, we are ready to start our optimization work!
Vectorized operations
Python is nice because it’s a relatively high level programming language, i.e. a lot of details are taken care of for you which makes programming faster, however the drawback is that you don’t always know what happens behind the scene.
For quite some time I have been using vanilla lists, not really understanding what exactly numpy arrays were designed for.
Finally came the day were I tried to perform simple operations on my 8+ millions rows dataset. When it takes about 10 seconds to execute a simple command, it may still seem acceptable until you try to run tens of these commands one after the other!
And then I tried to perform more complex operations, and I finally hit “out of memory” issues.
This is where numpy comes in handy. Numpy has been optimized to run vectorized operations, i.e. apply them to a whole array at a time, as opposed to applying the operation to each element of the array separately. And it makes a whole lot difference!
Consider adding 1 to each value of a list. We will reuse our list of one million element from earlier. If you do that manually:
%timeit [x+i for x in listy]
#1 loop, best of 3: 204 ms per loop
Now look at the vectorized version of this code:
%timeit listy+1
#1000 loops, best of 3: 1.02 ms per loop
Yes that’s right, 1.02 ms for vectorized operation versus 204 ms for non vectorized. 200 times faster!
How do you know when you perform vectorized versus non vectorized? From my experience, if you manipulate numpy objects (or pandas, which is build on top of numpy) and don’t use any loop (“for”, “while”…) or access subelements individually then you are probably using vectorized operations. Loops are killing you performance!
Also usually when you write a function and pass a whole array, series or dataframe to it, you’re on the right track. But if it takes as argument just one element, or a slice (such as a row), and you call it using apply on your dataframe/array, then expect poorer performances.
It can get pretty cumbersome to write vectorized operations… but when performance is at stake, and sometimes even the simple fact to complete your command, than you have no choice.
See in appendix two cases I had to face, one relatively simple, and the other one more complicated.
Cython
Cython is a way to compile part of your code in C in order to accelerate its execution, or even to embed C-like code in python.
I have been playing with cython in order to build various functions that beat numpy performance… And it’s a lot of fun!
I present below the cumulative sum example, where I managed to beat numpy. However this is not always easy… try beating numpy at quicksort… good luck with that!
Cumulative sum example
Let’s study the performance difference for several implementations of the cumulative sum function, i.e we want a function a function that computes the cumulative sums of consecutive elements in a given numeric “x”, i.e., a vector “r” with
Vanilla python
import numpy as np
def cumSum1(listy):
""" This function computes the cumulative sum of elements of a list"""
lengthy = len(listy)
listz = np.empty(lengthy)
listz[0] = listy[0]
for i in np.arange(1,lengthy):
listz[i] = listy[i]+listz[i-1]
return(listz)
-
Python, compiled in C
We only add the “%%cython” command at the beginning
%%cython
import numpy as np
def cumSum2(listy):
""" This function computes the cumulative sum of elements of a list"""
lengthy = len(listy)
listz = np.empty(lengthy)
listz[0] = listy[0]
for i in np.arange(1,lengthy):
listz[i] = listy[i]+listz[i-1]
return(listz)
-
Cython
This time we have a bit more work to do. All variables must be defined with proper types using the cdef command, and in order to optimize the code the “for” loop has been replaced by a “while” loop. Apart from that there is little difference with the previous version of the code.
%%cython
import numpy as np
cimport numpy as np
cpdef cumSum3(np.ndarray[np.int64_t] listy):
""" This function computes the cumulative sum of elements of a list"""
cdef int lengthy = listy.size
cdef int i = 1
cdef np.ndarray[np.double_t] listz = np.empty(lengthy)
listz[0] = listy[0]
cdef int previous = listy[0]
cdef int current = 0
while i < lengthy:
current = listy[i]+previous
listz[i] = current
previous = current
i += 1
return(listz)
I won’t go into more details of cython code here as pretty much by copying this example and others in appendix, you can get started. More on cython.org
Numpy
Numpy has a built-in “cumsum” function so we don’t need to write any code here.
Performance comparison
l = np.random.randint(1,100,10**7)
%timeit cumSum1(l)
%timeit cumSum2(l)
%timeit cumSum3(l)
%timeit np.cumsum(l)
#1 loop, best of 3: 20 s per loop
#1 loop, best of 3: 19.8 s per loop
#10 loops, best of 3: 20.8 ms per loop
#10 loops, best of 3: 34 ms per loop
And the winner is… our cython function cumSum3 !!!
Whilst there is little difference between vanilla and compiled (I have seen sometimes more impressive gains), the move to cython simply drives a drastic improvement of nearly 1000 times!
And numpy runs 70% slower than our cython function.
Checking the actual C code
When running the %%cython command, you can add a “-a” flag that shows the C code generated for each line of cython. For instance:
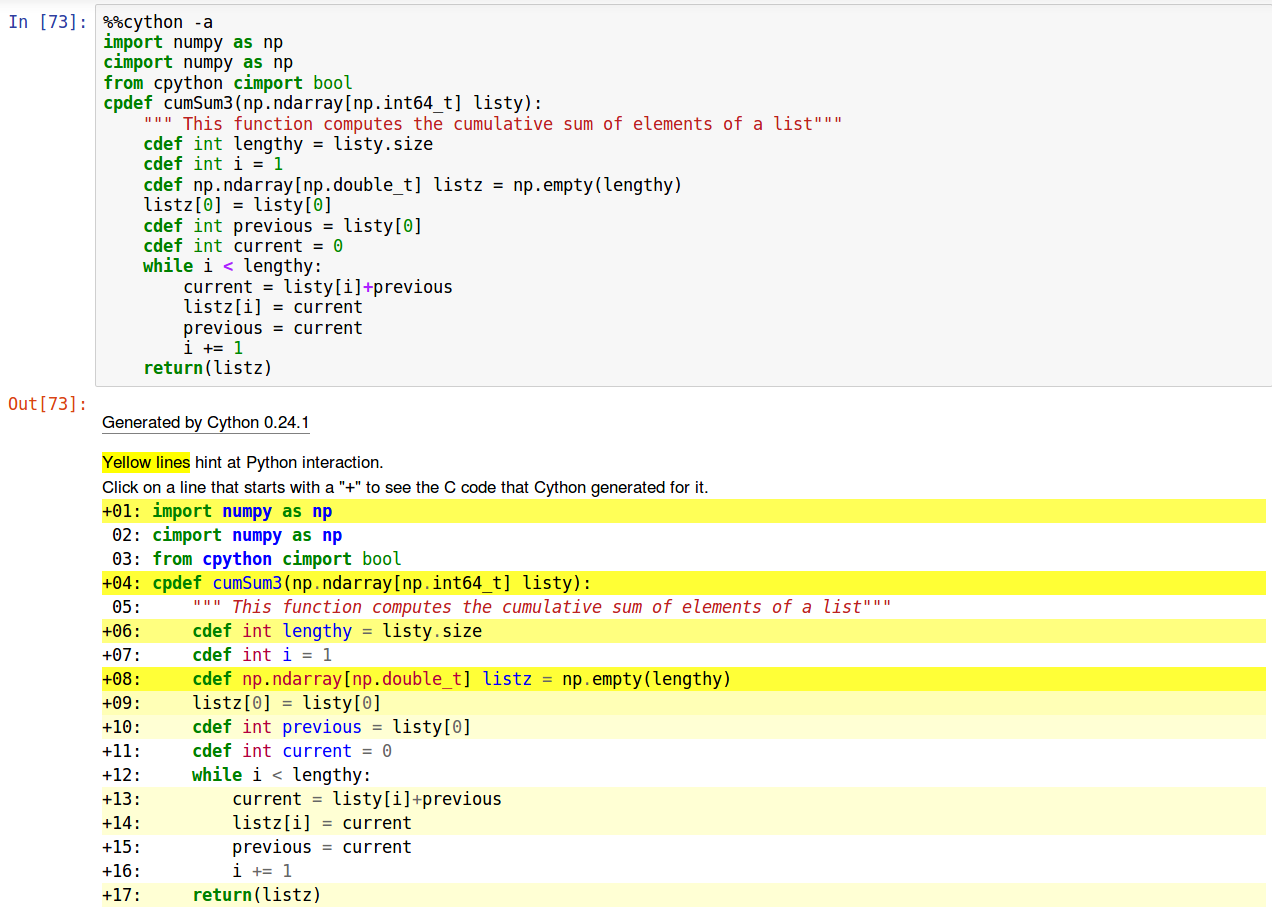
If lines are highlighted in yellow it means they lead to many lines of code (high complexity) and if this code is repeated many times (embedded in a loop), it’s a sign you may want to optimze it.
This tool is great to spot what part of your code are not optimal and need enhancement. For instance, if you write this same code with a “for” loop instead of “while”, you can see that it’s translated into many more lines of C code (dark yellow highlight):
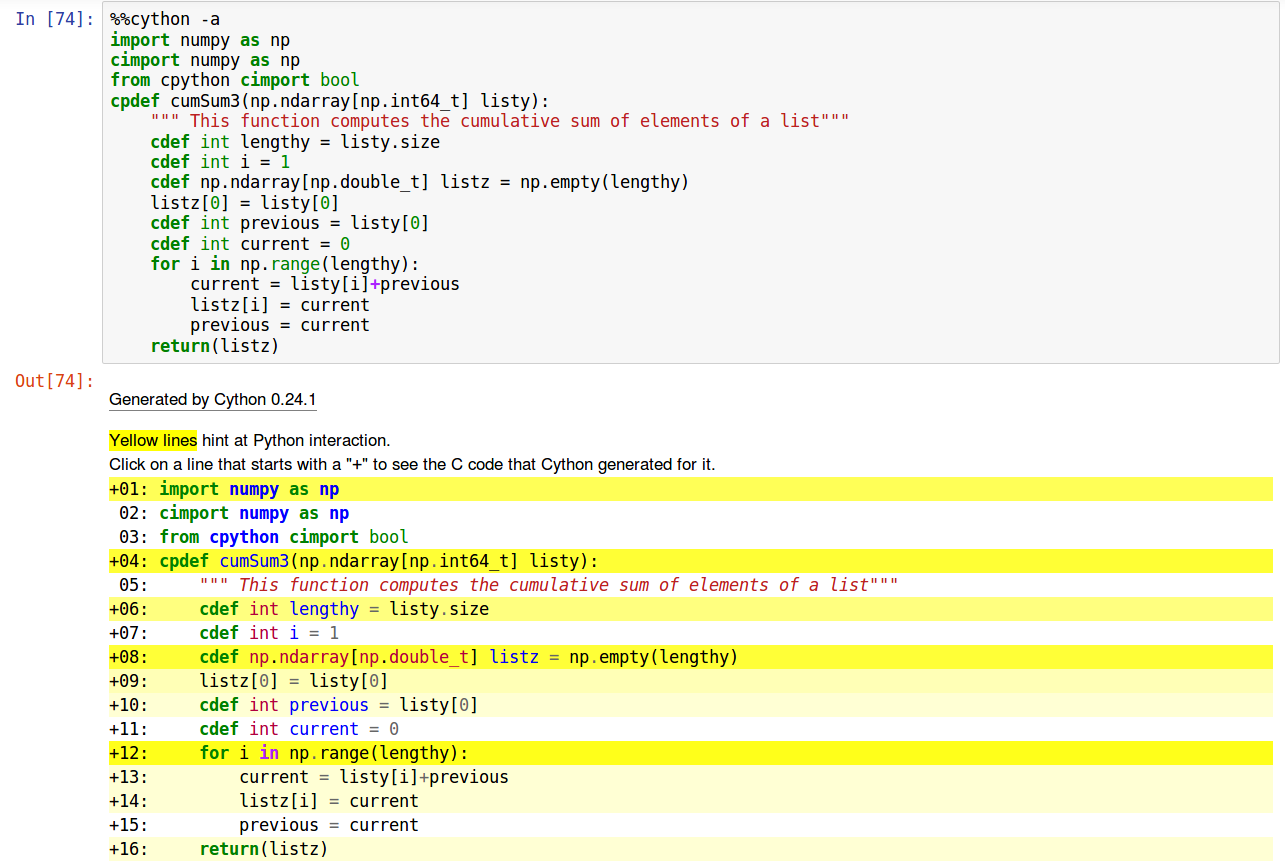
In that case the difference is probably small, but if you start embedding “for” loops, it will drive performance degradation, much more than embedded “while” loop.
This is one example amongst many. I have not finished exploring all the optimization opportunities, and I encourage you to try on your own!
Conclusion
This blog post showed two relatively easy ways to optimze python code. Undoubtly there are other tehniques, and in some cases data is so large that it won’t fit in memory, in which case you need to move to distributed processing, with spark for instance.
However these two techniques are a must know for anyone serious about writting fast and efficient code.
Enjoy!
Appendix
In this appendix I provide a few more examples of code optimization: 2 with vectorized operations, 2 with cython.
Vectorized example 1
In this case I simply wanted to map categorical values to numbers.
Non vectorized
gender_dict = {
'Male':0,
'Female':1,
'UNDEFINED':np.nan
}
%timeit df["Gender"].apply(lambda x: gender_dict[x])
#1 loop, best of 3: 2.65 s per loop
Vectorires
def set_gender(col):
res = pd.Series(['MALE' for _ in range(col.size)])
res = np.where(col == "UNDEFINED",np.nan,res)
res = np.where(col == "Female",1,res)
return res
%timeit set_gender(df["Gender"])
#1 loop, best of 3: 2.04 s per loop
The time difference may not seem so impressive here, but it’s already 30% less. And in fact other more complicated examples of the same type (with more categories for instance) have shown a much greater improvement by using vectorization.
Vectorized example 2
My data set contains 8+ million purchase records. I wanted to calculate how many days took place between each purchase, and put that information in a new column. In order to perform this as a vector operation, then only way I found was to duplicate the customer id (index) and purchase dates, and to shift them by one row. As a result you can compare dates from the original date column and from the new date column on the same row, and therefore no loop or complicated individual element access is required!
def days_between_purchases(df):
# Create a shifted copy of indices (=customer ids)
indices = df["index"]
first = indices[0]
new_indices = indices[1:]
new_indices[indices.size] = first
new_indices.index = indices.index
# Create a shifted copy of purchase dates
dates = df["Date of purchase"]
first = dates[0]
new_dates = dates[1:]
new_dates[dates.size] = first
new_dates.index = dates.index
# Add date when customer match
res = np.where((indices == new_indices),np.floor((new_dates - dates).astype('timedelta64[D]')),np.NaN)
return res
Without vectorization, creating the new row was simply craashing my jupyter notebook. With vectorization, it’s executed within a fraction of a second.
Cython example 1: is a list a palindrome
Python compiled in C
%%cython
def palindrome(listy):
res = True
i=0
while True & (i<=len(listy)//2):
res = res & (listy[i] == listy[-i-1])
i+=1
return res
Cython
%%cython
import numpy as np
cimport numpy as np
from cpython cimport bool
cpdef cpalindrome(np.ndarray[np.int64_t] listy):
cdef bool res = True
cdef int i = 0
cdef int lengthy = listy.size//2
cdef int remainder = listy.size%2
cdef np.ndarray[np.int64_t] list1 = listy[:lengthy]
cdef np.ndarray[np.int64_t] list2 = listy[remainder+lengthy:]
list2 = list2[::-1]
return np.array_equal(list1,list2)
Results
l1 = np.random.randint(1,100,10**7)
l2 = l1[::-1]
l = np.append(l1,l2)
%timeit palindrome(l)
%timeit cpalindrome(l)
#1 loop, best of 3: 2.63 s per loop
#100 loops, best of 3: 13.6 ms per loop
-
Cython example 2: reverse elements of a list
Python compiled in C
%%cython
import numpy as np
def invert(listy):
""" This function reverses the order of elements of a list"""
lengthy = len(listy)
listz = np.empty(lengthy)
for i in np.arange(lengthy):
listz[-i-1] = listy[i]
return(listz)
Cython
%%cython
import numpy as np
cimport numpy as np
from cpython cimport bool
cpdef cinvert(np.ndarray[np.int64_t] listy):
""" This function reverses the order of elements of a list"""
cdef int lengthy = listy.size
cdef int i = 0
cdef int j = lengthy-1
cdef np.ndarray[np.double_t] listz = np.empty(lengthy)
while i<lengthy:
listz[j] = listy[i]
i += 1
j -= 1
return(listz)
Results
l = np.random.randint(1,100,10**7)
%timeit invert(l)
%timeit cinvert(l)
#1 loop, best of 3: 4.42 s per loop
#10 loops, best of 3: 25.7 ms per loop